Reference maping using scGen#
In this tutorial, we are going to build a reference atlas using scGen and also map two new query datasets on the top of the reference atlas.
Note: scGen requires cell-type labels for data integration. The method outputs both corrected gene expression and also latent space.
[1]:
import os
import sys
sys.path.insert(0, "../")
import warnings
warnings.simplefilter(action='ignore', category=FutureWarning)
warnings.simplefilter(action='ignore', category=UserWarning)
[2]:
import scanpy as sc
import scarches as sca
from scarches.dataset.trvae.data_handling import remove_sparsity
import matplotlib.pyplot as plt
import numpy as np
import gdown
[3]:
sc.settings.set_figure_params(dpi=200, frameon=False)
sc.set_figure_params(dpi=200)
sc.set_figure_params(figsize=(4, 4))
set relevant anndata.obs labels and training hyperparameters#
[4]:
condition_key = 'study'
cell_type_key = 'cell_type'
target_conditions = ['Pancreas CelSeq2', 'Pancreas SS2']
epoch = 50
early_stopping_kwargs = {
"early_stopping_metric": "val_loss",
"patience": 20,
"threshold": 0,
"reduce_lr": True,
"lr_patience": 13,
"lr_factor": 0.1,
}
Download Dataset and split into reference dataset and query dataset#
[5]:
url = 'https://drive.google.com/uc?id=1ehxgfHTsMZXy6YzlFKGJOsBKQ5rrvMnd'
output = 'pancreas.h5ad'
gdown.download(url, output, quiet=False)
Downloading...
From: https://drive.google.com/uc?id=1ehxgfHTsMZXy6YzlFKGJOsBKQ5rrvMnd
To: /home/mo/projects/scarches/notebooks/pancreas.h5ad
126MB [00:01, 102MB/s]
[5]:
'pancreas.h5ad'
Imortant note : scGen requires normalized and log-transformed data in ``adata.X``
[6]:
adata = sc.read('pancreas.h5ad')
original uncorrected data UMAP#
[7]:
sc.pp.neighbors(adata)
WARNING: You’re trying to run this on 1000 dimensions of `.X`, if you really want this, set `use_rep='X'`.
Falling back to preprocessing with `sc.pp.pca` and default params.
[8]:
sc.tl.umap(adata)
[9]:
sc.pl.umap(adata, color=['study', 'cell_type'],
frameon=False, wspace=0.6)
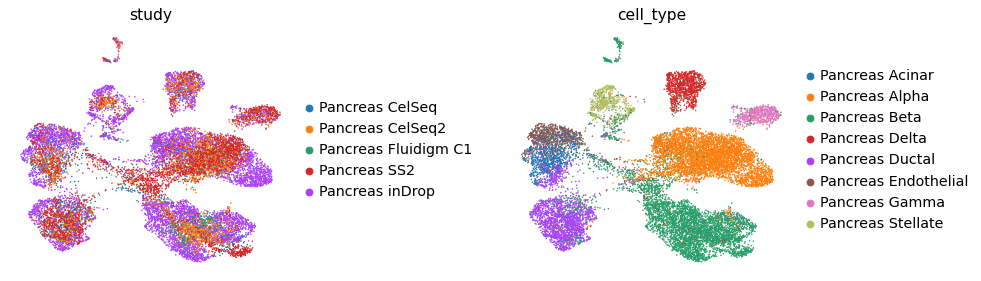
As observed above these Pancreas studies are seperated accroding to source of study
Here we use the CelSeq2 and SS2 studies as query data and the other 3 studies to build as reference atlas.
[10]:
adata = remove_sparsity(adata) # remove sparsity
source_adata = adata[~adata.obs[condition_key].isin(target_conditions)].copy()
target_adata = adata[adata.obs[condition_key].isin(target_conditions)].copy()
Create scGen model and train it on reference dataset#
Create the scgen model instance
[11]:
network = sca.models.scgen(adata = source_adata,
hidden_layer_sizes=[256,128])
INITIALIZING NEW NETWORK..............
Encoder Architecture:
Input Layer in, out: 1000 256
Hidden Layer 1 in/out: 256 128
Mean/Var Layer in/out: 128 10
Decoder Architecture:
First Layer in, out 10 128
Hidden Layer 1 in/out: 128 256
Output Layer in/out: 256 1000
[12]:
network.train(n_epochs=epoch, early_stopping_kwargs = early_stopping_kwargs)
|████████████████████| 100.0% - epoch_loss: 1.9351395184 - val_loss: 1.8963928728
Saving best state of network...
Best State was in Epoch 87
Correct batches in reference data#
This function returns corrected gene expression in adata.X, raw uncorrected data in adata.obsm["original_data"]. Also it returns uncorrected data in adata.layers["original_data"].
The low-dimensional corrected latent space in adata.obsm["latent_corrected"]
[13]:
corrected_reference_adata = network.batch_removal(source_adata, batch_key="study", cell_label_key="cell_type",return_latent=True)
Corrected gene expression
[14]:
sc.pp.neighbors(corrected_reference_adata)
sc.tl.umap(corrected_reference_adata)
sc.pl.umap(corrected_reference_adata, color=["study", "cell_type"], wspace=.5, frameon=False)
WARNING: You’re trying to run this on 1000 dimensions of `.X`, if you really want this, set `use_rep='X'`.
Falling back to preprocessing with `sc.pp.pca` and default params.
... storing 'cell_type' as categorical
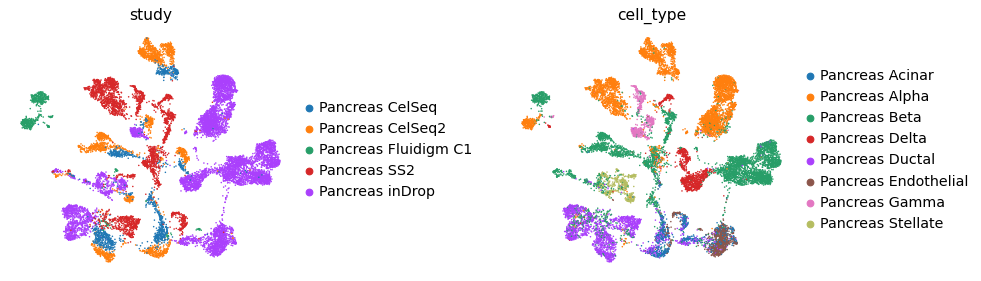
We an also use low-dim corrected reference data
[15]:
sc.pp.neighbors(corrected_reference_adata, use_rep="latent_corrected")
sc.tl.umap(corrected_reference_adata)
sc.pl.umap(corrected_reference_adata,
color=['study', 'cell_type'],
frameon=False,
wspace=0.6,
)
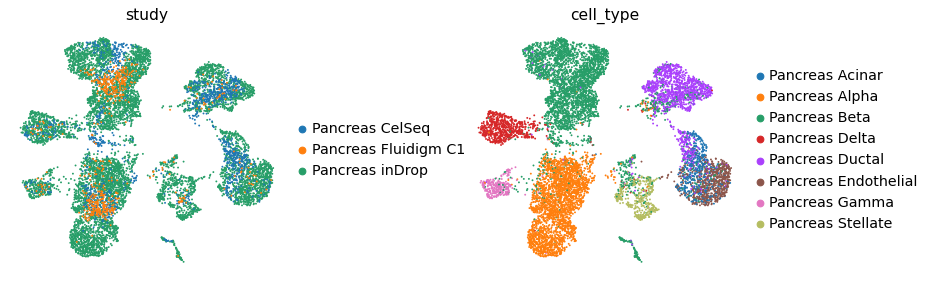
After training, the model can be saved for later use and projection of new query studies
[16]:
ref_path = './ref_model/'
network.save(ref_path, overwrite=True)
[17]:
os.getcwd()
[17]:
'/home/mo/projects/scarches/notebooks'
Project query on top of the reference#
query data needs to be preprocessed same way as reference data with same genes
This function need pretrained reference model, corrected gene expression from reference data and incorrected query data
[18]:
# here we pass the saved model from a file to the map query
integrated_query = sca.models.scgen.map_query_data(reference_model = network,
corrected_reference = corrected_reference_adata,
query = target_adata,
batch_key = 'study',
return_latent=True)
INITIALIZING NEW NETWORK..............
Encoder Architecture:
Input Layer in, out: 1000 256
Hidden Layer 1 in/out: 256 128
Mean/Var Layer in/out: 128 10
Decoder Architecture:
First Layer in, out 10 128
Hidden Layer 1 in/out: 128 256
Output Layer in/out: 256 1000
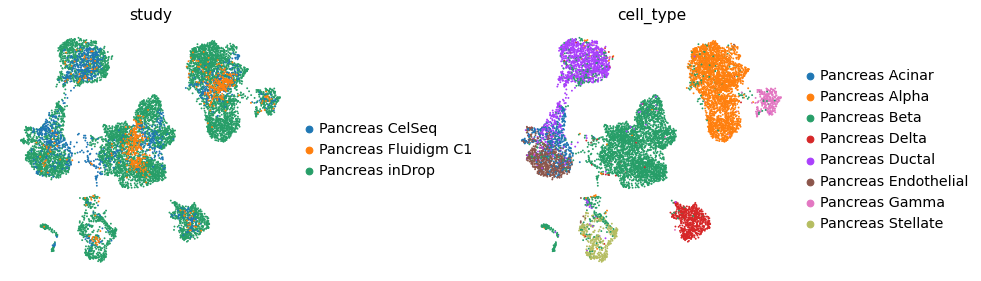
Plot the latent space of integrated query and reference#
[19]:
sc.pp.neighbors(integrated_query, use_rep="latent_corrected")
sc.tl.umap(integrated_query)
sc.pl.umap(
integrated_query,
color=["study", "cell_type"],
frameon=False,
wspace=0.6)
... storing 'batch' as categorical
... storing 'study' as categorical
... storing 'cell_type' as categorical
... storing 'original_batch' as categorical

Plot corrected gene expression space of integrated query and reference#
[20]:
sc.pp.neighbors(integrated_query)
sc.tl.umap(integrated_query)
sc.pl.umap(
integrated_query,
color=["study", "cell_type"],
frameon=False,
wspace=0.6)
WARNING: You’re trying to run this on 1000 dimensions of `.X`, if you really want this, set `use_rep='X'`.
Falling back to preprocessing with `sc.pp.pca` and default params.